3D examples¶
These examples require raw data which are automatically
downloaded from the source repository by the script
example_helper.py.
Please make sure that this script is present in the example
script folder.
Note
Windows users:
If you cannot run these examples directly, because you are getting
a RuntimeError with the text “An attempt has been made to start
a new process before the current process has finished its
bootstrapping phase”, please add the line
if __name__ == "__main__": at the beginning of the file
and indent the rest of the file by four spaces, i.e.
if __name__ == "__main__":
import matplotlib.pylab as plt
import nrefocus
import numpy as np
import odtbrain as odt
# etc.
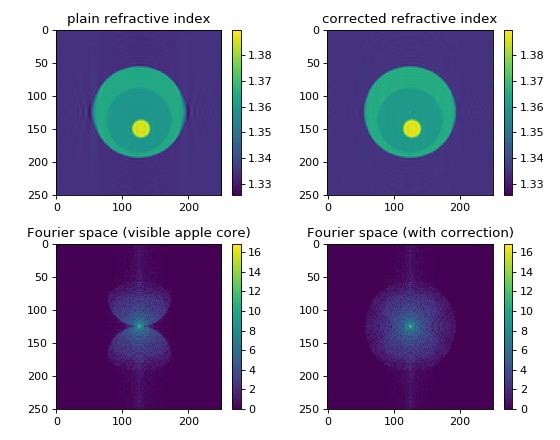
Missing apple core correction¶
The missing apple core [VDYH09] is a phenomenon in diffraction tomography that is a result of the fact the the Fourier space is not filled completely when the sample is rotated only about a single axis. The resulting artifacts include ringing and blurring in the reconstruction parallel to the original rotation axis. By enforcing constraints (refractive index real-valued and larger than the surrounding medium), these artifacts can be attenuated.
This example generates an artificial sinogram using the Python library cellsino (The example parameters are reused from this example). The sinogram is then reconstructed with the backpropagation algorithm and the missing apple core correction is applied.
Note
The missing apple core correction odtbrain.apple.correct()
was implemented in version 0.3.0 and is thus not used in the
older examples.
backprop_from_rytov_3d_phantom_apple.py
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 | import matplotlib.pylab as plt
import numpy as np
import cellsino
import odtbrain as odt
# number of sinogram angles
num_ang = 160
# sinogram acquisition angles
angles = np.linspace(0, 2*np.pi, num_ang, endpoint=False)
# detector grid size
grid_size = (250, 250)
# vacuum wavelength [m]
wavelength = 550e-9
# pixel size [m]
pixel_size = 0.08e-6
# refractive index of the surrounding medium
medium_index = 1.335
# initialize cell phantom
phantom = cellsino.phantoms.SimpleCell()
# initialize sinogram with geometric parameters
sino = cellsino.Sinogram(phantom=phantom,
wavelength=wavelength,
pixel_size=pixel_size,
grid_size=grid_size)
# compute sinogram (field according to Rytov approximation and fluorescence)
sino = sino.compute(angles=angles, propagator="rytov", mode="field")
# reconstruction of refractive index
sino_rytov = odt.sinogram_as_rytov(sino)
f = odt.backpropagate_3d(uSin=sino_rytov,
angles=angles,
res=wavelength/pixel_size,
nm=medium_index)
ri = odt.odt_to_ri(f=f,
res=wavelength/pixel_size,
nm=medium_index)
# apple core correction
fc = odt.apple.correct(f=f,
res=wavelength/pixel_size,
nm=medium_index,
method="sh")
ric = odt.odt_to_ri(f=fc,
res=wavelength/pixel_size,
nm=medium_index)
# plotting
idx = ri.shape[2] // 2
# log-scaled power spectra
ft = np.log(1 + np.abs(np.fft.fftshift(np.fft.fftn(ri))))
ftc = np.log(1 + np.abs(np.fft.fftshift(np.fft.fftn(ric))))
plt.figure(figsize=(7, 5.5))
plotkwri = {"vmax": ri.real.max(),
"vmin": ri.real.min(),
"interpolation": "none",
}
plotkwft = {"vmax": ft.max(),
"vmin": 0,
"interpolation": "none",
}
ax1 = plt.subplot(221, title="plain refractive index")
mapper = ax1.imshow(ri[:, :, idx].real, **plotkwri)
plt.colorbar(mappable=mapper, ax=ax1)
ax2 = plt.subplot(222, title="corrected refractive index")
mapper = ax2.imshow(ric[:, :, idx].real, **plotkwri)
plt.colorbar(mappable=mapper, ax=ax2)
ax3 = plt.subplot(223, title="Fourier space (visible apple core)")
mapper = ax3.imshow(ft[:, :, idx], **plotkwft)
plt.colorbar(mappable=mapper, ax=ax3)
ax4 = plt.subplot(224, title="Fourier space (with correction)")
mapper = ax4.imshow(ftc[:, :, idx], **plotkwft)
plt.colorbar(mappable=mapper, ax=ax4)
plt.tight_layout()
plt.show()
|
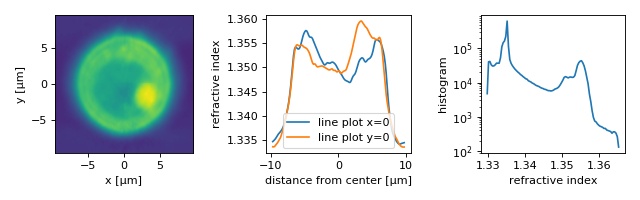
HL60 cell¶
The quantitative phase data of an HL60 S/4 cell were recorded using QLSI. The original dataset was used in a previous publication [SCG+17] to illustrate the capabilities of combined fluorescence and refractive index tomography.
The example data set is already aligned and background-corrected as described in the original publication and the fluorescence data are not included. The lzma-archive contains the sinogram data stored in the qpimage file format and the rotational positions of each sinogram image as a text file.
The figure reproduces parts of figure 4 of the original manuscript. Note that minor deviations from the original figure can be attributed to the strong compression (scale offset filter) and due to the fact that the original sinogram images were cropped from 196x196 px to 140x140 px (which in particular affects the background-part of the refractive index histogram).
The raw data is available on figshare <https://doi.org/10.6084/m9.figshare.8055407.v1> (hl60_sinogram_qpi.h5).
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 | import pathlib
import tarfile
import tempfile
import matplotlib.pylab as plt
import numpy as np
import odtbrain as odt
import qpimage
from example_helper import get_file, extract_lzma
# ascertain the data
path = get_file("qlsi_3d_hl60-cell_A140.tar.lzma")
tarf = extract_lzma(path)
tdir = tempfile.mkdtemp(prefix="odtbrain_example_")
with tarfile.open(tarf) as tf:
tf.extract("series.h5", path=tdir)
angles = np.loadtxt(tf.extractfile("angles.txt"))
# extract the complex field sinogram from the qpimage series data
h5file = pathlib.Path(tdir) / "series.h5"
with qpimage.QPSeries(h5file=h5file, h5mode="r") as qps:
qp0 = qps[0]
meta = qp0.meta
sino = np.zeros((len(qps), qp0.shape[0], qp0.shape[1]), dtype=np.complex)
for ii in range(len(qps)):
sino[ii] = qps[ii].field
# perform backgpropagation
u_sinR = odt.sinogram_as_rytov(sino)
res = meta["wavelength"] / meta["pixel size"]
nm = meta["medium index"]
fR = odt.backpropagate_3d(uSin=u_sinR,
angles=angles,
res=res,
nm=nm)
ri = odt.odt_to_ri(fR, res, nm)
# plot results
ext = meta["pixel size"] * 1e6 * 70
kw = {"vmin": ri.real.min(),
"vmax": ri.real.max(),
"extent": [-ext, ext, -ext, ext]}
fig, axes = plt.subplots(1, 3, figsize=(8, 2.5))
axes[0].imshow(ri[70, :, :].real, **kw)
axes[0].set_xlabel("x [µm]")
axes[0].set_ylabel("y [µm]")
x = np.linspace(-ext, ext, 140)
axes[1].plot(x, ri[70, :, 70], label="line plot x=0")
axes[1].plot(x, ri[70, 70, :], label="line plot y=0")
axes[1].set_xlabel("distance from center [µm]")
axes[1].set_ylabel("refractive index")
axes[1].legend()
hist, xh = np.histogram(ri.real, bins=100)
axes[2].plot(xh[1:], hist)
axes[2].set_yscale('log')
axes[2].set_xlabel("refractive index")
axes[2].set_ylabel("histogram")
plt.tight_layout()
plt.show()
|
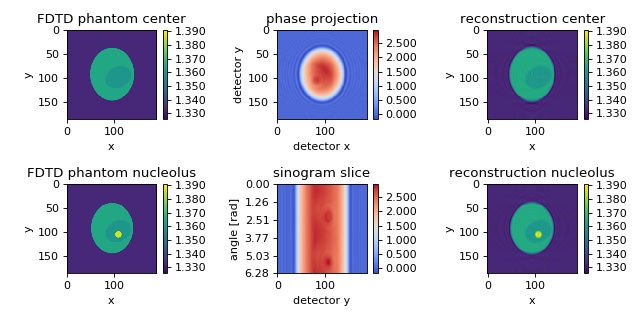
FDTD cell phantom¶
The in silico data set was created with the FDTD software meep. The data are 2D projections of a 3D refractive index phantom. The reconstruction of the refractive index with the Rytov approximation is in good agreement with the phantom that was used in the simulation. The data are downsampled by a factor of two. The rotational axis is the y-axis. A total of 180 projections are used for the reconstruction. A detailed description of this phantom is given in [MSG15a].
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 | import matplotlib.pylab as plt
import numpy as np
import odtbrain as odt
from example_helper import load_data
sino, angles, phantom, cfg = \
load_data("fdtd_3d_sino_A180_R6.500.tar.lzma")
A = angles.shape[0]
print("Example: Backpropagation from 3D FDTD simulations")
print("Refractive index of medium:", cfg["nm"])
print("Measurement position from object center:", cfg["lD"])
print("Wavelength sampling:", cfg["res"])
print("Number of projections:", A)
print("Performing backpropagation.")
# Apply the Rytov approximation
sinoRytov = odt.sinogram_as_rytov(sino)
# perform backpropagation to obtain object function f
f = odt.backpropagate_3d(uSin=sinoRytov,
angles=angles,
res=cfg["res"],
nm=cfg["nm"],
lD=cfg["lD"]
)
# compute refractive index n from object function
n = odt.odt_to_ri(f, res=cfg["res"], nm=cfg["nm"])
sx, sy, sz = n.shape
px, py, pz = phantom.shape
sino_phase = np.angle(sino)
# compare phantom and reconstruction in plot
fig, axes = plt.subplots(2, 3, figsize=(8, 4))
kwri = {"vmin": n.real.min(), "vmax": n.real.max()}
kwph = {"vmin": sino_phase.min(), "vmax": sino_phase.max(),
"cmap": "coolwarm"}
# Phantom
axes[0, 0].set_title("FDTD phantom center")
rimap = axes[0, 0].imshow(phantom[px // 2], **kwri)
axes[0, 0].set_xlabel("x")
axes[0, 0].set_ylabel("y")
axes[1, 0].set_title("FDTD phantom nucleolus")
axes[1, 0].imshow(phantom[int(px / 2 + 2 * cfg["res"])], **kwri)
axes[1, 0].set_xlabel("x")
axes[1, 0].set_ylabel("y")
# Sinogram
axes[0, 1].set_title("phase projection")
phmap = axes[0, 1].imshow(sino_phase[A // 2, :, :], **kwph)
axes[0, 1].set_xlabel("detector x")
axes[0, 1].set_ylabel("detector y")
axes[1, 1].set_title("sinogram slice")
axes[1, 1].imshow(sino_phase[:, :, sino.shape[2] // 2],
aspect=sino.shape[1] / sino.shape[0], **kwph)
axes[1, 1].set_xlabel("detector y")
axes[1, 1].set_ylabel("angle [rad]")
# set y ticks for sinogram
labels = np.linspace(0, 2 * np.pi, len(axes[1, 1].get_yticks()))
labels = ["{:.2f}".format(i) for i in labels]
axes[1, 1].set_yticks(np.linspace(0, len(angles), len(labels)))
axes[1, 1].set_yticklabels(labels)
axes[0, 2].set_title("reconstruction center")
axes[0, 2].imshow(n[sx // 2].real, **kwri)
axes[0, 2].set_xlabel("x")
axes[0, 2].set_ylabel("y")
axes[1, 2].set_title("reconstruction nucleolus")
axes[1, 2].imshow(n[int(sx / 2 + 2 * cfg["res"])].real, **kwri)
axes[1, 2].set_xlabel("x")
axes[1, 2].set_ylabel("y")
# color bars
cbkwargs = {"fraction": 0.045,
"format": "%.3f"}
plt.colorbar(phmap, ax=axes[0, 1], **cbkwargs)
plt.colorbar(phmap, ax=axes[1, 1], **cbkwargs)
plt.colorbar(rimap, ax=axes[0, 0], **cbkwargs)
plt.colorbar(rimap, ax=axes[1, 0], **cbkwargs)
plt.colorbar(rimap, ax=axes[0, 2], **cbkwargs)
plt.colorbar(rimap, ax=axes[1, 2], **cbkwargs)
plt.tight_layout()
plt.show()
|
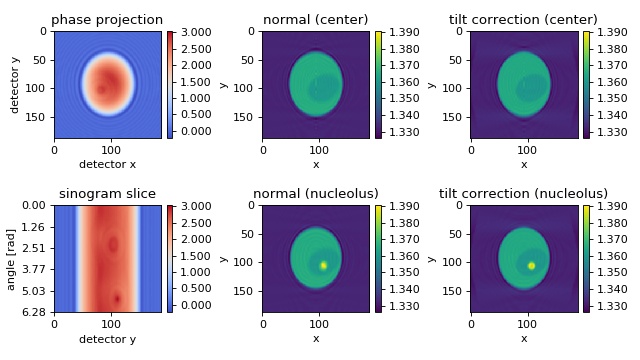
FDTD cell phantom with tilted axis of rotation¶
The in silico data set was created with the
FDTD software meep. The data
are 2D projections of a 3D refractive index phantom that is rotated
about an axis which is tilted by 0.2 rad (11.5 degrees) with respect
to the imaging plane. The example showcases the method
odtbrain.backpropagate_3d_tilted() which takes into account
such a tilted axis of rotation. The data are downsampled by a factor
of two. A total of 220 projections are used for the reconstruction.
Note that the information required for reconstruction decreases as the
tilt angle increases. If the tilt angle is 90 degrees w.r.t. the
imaging plane, then we get a rotating image of a cell (not images of a
rotating cell) and tomographic reconstruction is impossible. A brief
description of this algorithm is given in [MSCG15].
The first column shows the measured phase, visualizing the tilt (compare to other examples). The second column shows a reconstruction that does not take into account the tilted axis of rotation; the result is a blurry reconstruction. The third column shows the improved reconstruction; the known tilted axis of rotation is used in the reconstruction process.
backprop_from_fdtd_3d_tilted.py
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 | import matplotlib.pylab as plt
import numpy as np
import odtbrain as odt
from example_helper import load_data
sino, angles, phantom, cfg = \
load_data("fdtd_3d_sino_A220_R6.500_tiltyz0.2.tar.lzma")
A = angles.shape[0]
print("Example: Backpropagation from 3D FDTD simulations")
print("Refractive index of medium:", cfg["nm"])
print("Measurement position from object center:", cfg["lD"])
print("Wavelength sampling:", cfg["res"])
print("Axis tilt in y-z direction:", cfg["tilt_yz"])
print("Number of projections:", A)
print("Performing normal backpropagation.")
# Apply the Rytov approximation
sinoRytov = odt.sinogram_as_rytov(sino)
# Perform naive backpropagation
f_naiv = odt.backpropagate_3d(uSin=sinoRytov,
angles=angles,
res=cfg["res"],
nm=cfg["nm"],
lD=cfg["lD"]
)
print("Performing tilted backpropagation.")
# Determine tilted axis
tilted_axis = [0, np.cos(cfg["tilt_yz"]), np.sin(cfg["tilt_yz"])]
# Perform tilted backpropagation
f_tilt = odt.backpropagate_3d_tilted(uSin=sinoRytov,
angles=angles,
res=cfg["res"],
nm=cfg["nm"],
lD=cfg["lD"],
tilted_axis=tilted_axis,
)
# compute refractive index n from object function
n_naiv = odt.odt_to_ri(f_naiv, res=cfg["res"], nm=cfg["nm"])
n_tilt = odt.odt_to_ri(f_tilt, res=cfg["res"], nm=cfg["nm"])
sx, sy, sz = n_tilt.shape
px, py, pz = phantom.shape
sino_phase = np.angle(sino)
# compare phantom and reconstruction in plot
fig, axes = plt.subplots(2, 3, figsize=(8, 4.5))
kwri = {"vmin": n_tilt.real.min(), "vmax": n_tilt.real.max()}
kwph = {"vmin": sino_phase.min(), "vmax": sino_phase.max(),
"cmap": "coolwarm"}
# Sinogram
axes[0, 0].set_title("phase projection")
phmap = axes[0, 0].imshow(sino_phase[A // 2, :, :], **kwph)
axes[0, 0].set_xlabel("detector x")
axes[0, 0].set_ylabel("detector y")
axes[1, 0].set_title("sinogram slice")
axes[1, 0].imshow(sino_phase[:, :, sino.shape[2] // 2],
aspect=sino.shape[1] / sino.shape[0], **kwph)
axes[1, 0].set_xlabel("detector y")
axes[1, 0].set_ylabel("angle [rad]")
# set y ticks for sinogram
labels = np.linspace(0, 2 * np.pi, len(axes[1, 1].get_yticks()))
labels = ["{:.2f}".format(i) for i in labels]
axes[1, 0].set_yticks(np.linspace(0, len(angles), len(labels)))
axes[1, 0].set_yticklabels(labels)
axes[0, 1].set_title("normal (center)")
rimap = axes[0, 1].imshow(n_naiv[sx // 2].real, **kwri)
axes[0, 1].set_xlabel("x")
axes[0, 1].set_ylabel("y")
axes[1, 1].set_title("normal (nucleolus)")
axes[1, 1].imshow(n_naiv[int(sx / 2 + 2 * cfg["res"])].real, **kwri)
axes[1, 1].set_xlabel("x")
axes[1, 1].set_ylabel("y")
axes[0, 2].set_title("tilt correction (center)")
axes[0, 2].imshow(n_tilt[sx // 2].real, **kwri)
axes[0, 2].set_xlabel("x")
axes[0, 2].set_ylabel("y")
axes[1, 2].set_title("tilt correction (nucleolus)")
axes[1, 2].imshow(n_tilt[int(sx / 2 + 2 * cfg["res"])].real, **kwri)
axes[1, 2].set_xlabel("x")
axes[1, 2].set_ylabel("y")
# color bars
cbkwargs = {"fraction": 0.045,
"format": "%.3f"}
plt.colorbar(phmap, ax=axes[0, 0], **cbkwargs)
plt.colorbar(phmap, ax=axes[1, 0], **cbkwargs)
plt.colorbar(rimap, ax=axes[0, 1], **cbkwargs)
plt.colorbar(rimap, ax=axes[1, 1], **cbkwargs)
plt.colorbar(rimap, ax=axes[0, 2], **cbkwargs)
plt.colorbar(rimap, ax=axes[1, 2], **cbkwargs)
plt.tight_layout()
plt.show()
|
FDTD cell phantom with tilted and rolled axis of rotation¶
The in silico data set was created with the FDTD software meep. The data are 2D projections of a 3D refractive index phantom that is rotated about an axis which is tilted by 0.2 rad (11.5 degrees) with respect to the imaging plane and rolled by -.42 rad (-24.1 degrees) within the imaging plane. The data are the same as were used in the previous example. A brief description of this algorithm is given in [MSCG15].
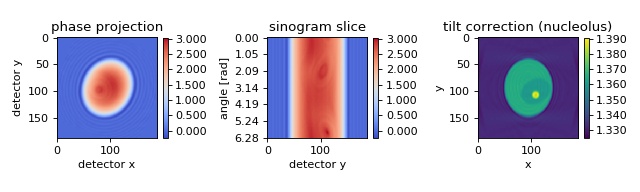
backprop_from_fdtd_3d_tilted2.py
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 | import matplotlib.pylab as plt
import numpy as np
from scipy.ndimage import rotate
import odtbrain as odt
from example_helper import load_data
sino, angles, phantom, cfg = \
load_data("fdtd_3d_sino_A220_R6.500_tiltyz0.2.tar.lzma")
# Perform titlt by -.42 rad in detector plane
rotang = -0.42
rotkwargs = {"mode": "constant",
"order": 2,
"reshape": False,
}
for ii in range(len(sino)):
sino[ii].real = rotate(
sino[ii].real, np.rad2deg(rotang), cval=1, **rotkwargs)
sino[ii].imag = rotate(
sino[ii].imag, np.rad2deg(rotang), cval=0, **rotkwargs)
A = angles.shape[0]
print("Example: Backpropagation from 3D FDTD simulations")
print("Refractive index of medium:", cfg["nm"])
print("Measurement position from object center:", cfg["lD"])
print("Wavelength sampling:", cfg["res"])
print("Axis tilt in y-z direction:", cfg["tilt_yz"])
print("Number of projections:", A)
# Apply the Rytov approximation
sinoRytov = odt.sinogram_as_rytov(sino)
# Determine tilted axis
tilted_axis = [0, np.cos(cfg["tilt_yz"]), np.sin(cfg["tilt_yz"])]
rotmat = np.array([
[np.cos(rotang), -np.sin(rotang), 0],
[np.sin(rotang), np.cos(rotang), 0],
[0, 0, 1],
])
tilted_axis = np.dot(rotmat, tilted_axis)
print("Performing tilted backpropagation.")
# Perform tilted backpropagation
f_tilt = odt.backpropagate_3d_tilted(uSin=sinoRytov,
angles=angles,
res=cfg["res"],
nm=cfg["nm"],
lD=cfg["lD"],
tilted_axis=tilted_axis,
)
# compute refractive index n from object function
n_tilt = odt.odt_to_ri(f_tilt, res=cfg["res"], nm=cfg["nm"])
sx, sy, sz = n_tilt.shape
px, py, pz = phantom.shape
sino_phase = np.angle(sino)
# compare phantom and reconstruction in plot
fig, axes = plt.subplots(1, 3, figsize=(8, 2.4))
kwri = {"vmin": n_tilt.real.min(), "vmax": n_tilt.real.max()}
kwph = {"vmin": sino_phase.min(), "vmax": sino_phase.max(),
"cmap": "coolwarm"}
# Sinogram
axes[0].set_title("phase projection")
phmap = axes[0].imshow(sino_phase[A // 2, :, :], **kwph)
axes[0].set_xlabel("detector x")
axes[0].set_ylabel("detector y")
axes[1].set_title("sinogram slice")
axes[1].imshow(sino_phase[:, :, sino.shape[2] // 2],
aspect=sino.shape[1] / sino.shape[0], **kwph)
axes[1].set_xlabel("detector y")
axes[1].set_ylabel("angle [rad]")
# set y ticks for sinogram
labels = np.linspace(0, 2 * np.pi, len(axes[1].get_yticks()))
labels = ["{:.2f}".format(i) for i in labels]
axes[1].set_yticks(np.linspace(0, len(angles), len(labels)))
axes[1].set_yticklabels(labels)
axes[2].set_title("tilt correction (nucleolus)")
rimap = axes[2].imshow(n_tilt[int(sx / 2 + 2 * cfg["res"])].real, **kwri)
axes[2].set_xlabel("x")
axes[2].set_ylabel("y")
# color bars
cbkwargs = {"fraction": 0.045,
"format": "%.3f"}
plt.colorbar(phmap, ax=axes[0], **cbkwargs)
plt.colorbar(phmap, ax=axes[1], **cbkwargs)
plt.colorbar(rimap, ax=axes[2], **cbkwargs)
plt.tight_layout()
plt.show()
|
Mie sphere¶
The in silico data set was created with the Mie calculation software GMM-field. The data consist of a two-dimensional projection of a sphere with radius \(R=14\lambda\), refractive index \(n_\mathrm{sph}=1.006\), embedded in a medium of refractive index \(n_\mathrm{med}=1.0\) onto a detector which is \(l_\mathrm{D} = 20\lambda\) away from the center of the sphere.
The package nrefocus must be used to numerically focus
the detected field prior to the 3D backpropagation with ODTbrain.
In odtbrain.backpropagate_3d(), the parameter lD must
be set to zero (\(l_\mathrm{D}=0\)).
The figure shows the 3D reconstruction from Mie simulations of a perfect sphere using 200 projections. Missing angle artifacts are visible along the \(y\)-axis due to the \(2\pi\)-only coverage in 3D Fourier space.
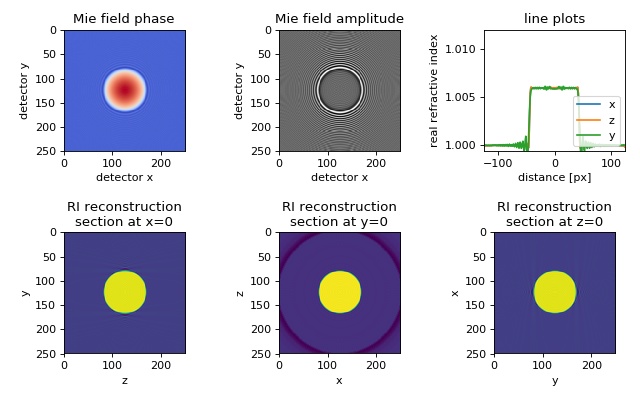
backprop_from_mie_3d_sphere.py
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 | import matplotlib.pylab as plt
import nrefocus
import numpy as np
import odtbrain as odt
from example_helper import load_data
Ex, cfg = load_data("mie_3d_sphere_field.zip",
f_sino_imag="mie_sphere_imag.txt",
f_sino_real="mie_sphere_real.txt",
f_info="mie_info.txt")
# Manually set number of angles:
A = 200
print("Example: Backpropagation from 3D Mie scattering")
print("Refractive index of medium:", cfg["nm"])
print("Measurement position from object center:", cfg["lD"])
print("Wavelength sampling:", cfg["res"])
print("Number of angles for reconstruction:", A)
print("Performing backpropagation.")
# Reconstruction angles
angles = np.linspace(0, 2 * np.pi, A, endpoint=False)
# Perform focusing
Ex = nrefocus.refocus(Ex,
d=-cfg["lD"]*cfg["res"],
nm=cfg["nm"],
res=cfg["res"],
)
# Create sinogram
u_sin = np.tile(Ex.flat, A).reshape(A, int(cfg["size"]), int(cfg["size"]))
# Apply the Rytov approximation
u_sinR = odt.sinogram_as_rytov(u_sin)
# Backpropagation
fR = odt.backpropagate_3d(uSin=u_sinR,
angles=angles,
res=cfg["res"],
nm=cfg["nm"],
lD=0,
padfac=2.1,
save_memory=True)
# RI computation
nR = odt.odt_to_ri(fR, cfg["res"], cfg["nm"])
# Plotting
fig, axes = plt.subplots(2, 3, figsize=(8, 5))
axes = np.array(axes).flatten()
# field
axes[0].set_title("Mie field phase")
axes[0].set_xlabel("detector x")
axes[0].set_ylabel("detector y")
axes[0].imshow(np.angle(Ex), cmap="coolwarm")
axes[1].set_title("Mie field amplitude")
axes[1].set_xlabel("detector x")
axes[1].set_ylabel("detector y")
axes[1].imshow(np.abs(Ex), cmap="gray")
# line plot
axes[2].set_title("line plots")
axes[2].set_xlabel("distance [px]")
axes[2].set_ylabel("real refractive index")
center = int(cfg["size"] / 2)
x = np.arange(cfg["size"]) - center
axes[2].plot(x, nR[:, center, center].real, label="x")
axes[2].plot(x, nR[center, center, :].real, label="z")
axes[2].plot(x, nR[center, :, center].real, label="y")
axes[2].legend(loc=4)
axes[2].set_xlim((-center, center))
dn = abs(cfg["nsph"] - cfg["nm"])
axes[2].set_ylim((cfg["nm"] - dn / 10, cfg["nsph"] + dn))
axes[2].ticklabel_format(useOffset=False)
# cross sections
axes[3].set_title("RI reconstruction\nsection at x=0")
axes[3].set_xlabel("z")
axes[3].set_ylabel("y")
axes[3].imshow(nR[center, :, :].real)
axes[4].set_title("RI reconstruction\nsection at y=0")
axes[4].set_xlabel("x")
axes[4].set_ylabel("z")
axes[4].imshow(nR[:, center, :].real)
axes[5].set_title("RI reconstruction\nsection at z=0")
axes[5].set_xlabel("y")
axes[5].set_ylabel("x")
axes[5].imshow(nR[:, :, center].real)
plt.tight_layout()
plt.show()
|